Every litre of seawater is full of genetic material from all kinds of different organisms. Biologist Silke Laakmann and her team are pioneering techniques that use these DNA traces to determine the biodiversity of marine communities. This involves combining conventional methods of species identification with new genetic procedures.
The marine protected area of Sylt Outer Reef has unusually high biodiversity levels for North Sea standards: fields of hard substrates alternate with areas of gravel and coarse sand, interspersed with flat sandbanks. The stones provide a solid base for colourful reef communities of organisms such as frilled anenomes, mussels, bryozoans and sponges, while the sandbanks are home to various species of brittle stars and worms.
Numerous fish species thrive in these waters, among them cod, sprats and herring, as well as flatfish and rare European river lamprey. This rich food supply in turn attracts marine mammals such as harbour porpoises and seals.
Unsavory mixture with high information content
So far, assessing biodiversity in marine protected areas such as Sylt Outer Reef has been a very laborious process for environmental researchers: they cast nets of various mesh sizes to catch different sized marine species; use box corers to extract samples from the seafloor, and underwater cameras to take photos and make videos of the seabed.
But there are other ways to go about this: “All we need is a water sample,” says biologist Dr Silke Laakmann. The researcher and her team take advantage of the fact that a highly diluted collection of biological debris is just floating around in the sea: remnants of cells, mucous, scales, hairs, faeces and the remains of decaying organisms.
However unappetising this might sound, the genetic material this debris contains, when analysed correctly, can provide astonishing insights into the biodiversity of marine communities. The official term for this material is environmental DNA, or eDNA for short.
At the Helmholtz Institute for Functional Marine Biodiversity (HIFMB) at the University of Oldenburg, Laakmann is working towards the major goal of achieving reliable assessment of marine biodiversity levels using noth-ing but water samples. The HIFMB was founded in 2017 as a cooperation between the University of Oldenburg and the Alfred Wegener Institute, Helmholtz Centre for Polar and Marine Research.
eDNA: a powerful new tool
“We study whether the eDNA results correspond with reality – in other words, whether we actually register as many organisms as possible present in a given region,” says the researcher, who has been running the Focus Group Marine Molecular Ecology at the HIFMB since 2018. Two such focus groups are currently based at the institute. They are an instrument to provide targeted funding to promising mid career scientists and, at the same time, consolidate highly innovative research fields at the HIFMB.
The procedure at the heart of the work of Laakmann and her five-member team is also very much on trend. In recent years, eDNA analysis has become a powerful new tool in the environmental sciences. The method has huge potential as it allows analysing entire communities, however large, in one go, and provides all sorts of new insights into biodiversity. It also reduces environmental impact compared to traditional methods of sampling of marine organisms – an advantage which is particularly relevant in marine conservation areas.
Study object European oyster
In the collaborative CREATE project, headed by Oldenburg biodiversity expert and HIFMB Director Prof. Dr Helmut Hillebrand, eDNA plays a central role to analyse the biodiversity of marine protected areas as well as the connections between them. The European oyster is the project’s key species. “One key question is whether oyster larvae from the Borkum Reef Ground protected area move with the currents into other marine areas where they had not previously been restored,” explains Laakmann.
As part of the project, which is funded by the Federal Ministry of Education and Research in the framework of the German Marine Research Alliance, the biologist and her team currently compile an eDNA archive for the North Sea, aimed at documenting the current status of biodiversity and the effects of future environmental changes. Environmental scientist Dr Kingsly Chuo Beng, a postdoctoral researcher in the focus group and an expert in eDNA research, manages the corresponding work
package.
Change in biodiversity research
The researchers in Laakmann’s team have been laying the groundwork for this over the past four years: “Recently, a shift has taken place in biodiversity research, from what you might call the classical morphological methods of species identification to molecular ones,” the researcher explains.
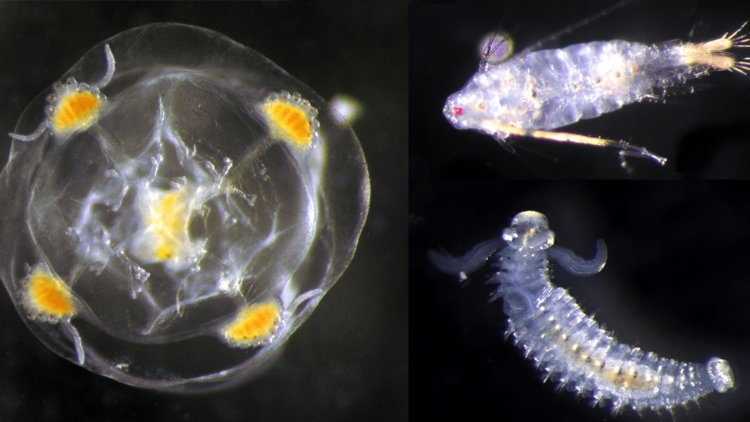
From microscope to genes, in other words. Her own training started in the conventional methods. “During my doctoral studies at the University of Bremen and my time as a postdoc at the Research Centre Senckenberg am Meer in Wilhelmshaven, I spent a lot of time in the sorting lab identifying organisms under the microscope according to their external form,” she recalls. Her speciality is zooplankton – aquatic organisms that are drifted along by currents in the oceans – in particular a diverse group of tiny crustaceans called copepods.
More than 3,000 samples collected
With the goal of establishing the novel genetic methods as a routine process for environmental monitoring, a comparison with conventional meth-ods is currently underway. Laakmann and her team focus primarily on zooplankton in the North Sea and the Baltic, but also analyse material from Patagonia, the Arctic and South Africa. They have collected more than 3,000 samples which will be preserved for further studies at the HIFMB.
To obtain an overview of biodiversity levels, the scientists generally use three different methods. They still collect plankton in the traditional way using long, thin nets. This often brings on board a brownish, flaky mass of sea organisms which can range in size from the head of a pin to a few millimetres.
“These samples are then splitted in two halves,” Lackmann says. One half goes to her and her team for conventional identification under a microscope or to semi-automated imaging analysis with Dr Astrid Cornils, a colleague at AWI. The other half is homogenised and then genetically analysed. The third method is eDNA analysis, whereby genetic material is extracted from water samples taken from various depths.
Gene segment COI is unique for each species
The molecular biology methods used for species identification are to some extent similar to those used for PCR tests in Covid test labs, in which characteristic areas of genes are amplified and sequenced for analysis.
The environmental sciences mostly concentrate on a gene segment that goes by the name of COI (cytochrome c oxidase subunit I). “The sequence of this special gene fragment is unique in every animal species,” Laakmann explains. In huge databases, researchers from various international initiatives inventory species using the genetic code of their COI gene.
Just as products in supermarkets are identified by barcodes on their packaging, every animal species ever registered in a database can be identified by its genetic code. As a PhD student and postdoc, Laakmann helped to expand this library of life by numerous species, among them organisms living in the water and on the seafloor of the North Sea, as well as copepods from the Arctic, the Antarctic and the deep sea.
I think it is important to make a connection with the organisms I study.
Silke Laakmann
When the vast assemblage of genetic material in an environmental sample is analysed in parallel – a process that gives rise to thousands of different DNA sequences – this is known as “metabarcoding”. “In our samples we have millions of these gene snippets from all sorts of organisms. After sequencing we compare the gene sequences with the database entries, so that in the end we have a list of species or groups,” Laakmann explains.
Over the past four years the researchers have developed a special toolkit for dealing with the eDNA. “We were interested in questions such as: How much water do we need in order to capture as many species as possible in one area? How often do we need to take samples? What should the filter look like? What databases should we use?” the biologist notes.
The team also studied threshold values and computational algorithms. Laakmann is satisfied with the results: “We now know that eDNA metabarcoding is indeed able to identify all the various groups of invertebrates and fish that live in the North Sea. And now we understand pretty good how best to apply the method to different questions.”
Adapting examinations to the environment
In order to examine whether or not particular species are present in large marine protected areas like Borkum Reef Ground or Sylt Outer Reef, it might make sense to take water samples from several different places, she says. In a study of bottom-dwelling organisms, the researchers are collaborating with a team from the AWI that carries out long-term observations in marine protected areas.
Here, they combine DNA analyses with sediment samples and underwater photos. In particularly dynamic marine areas, on the other hand, it may be necessary to take water samples at the same location over and over again for several days on end in order to record all species present.
Laakmann stresses the importance of not relying solely on the genetic method, even in the future. “I would like to use integrative methods, which means combining conventional and new methods. That delivers the best of both worlds.”
As a marine biologist, she wouldn’t want to miss out on identifying tiny amphipods, hydromedusae and opossum shrimps under the microscope. “I think it is important so I can make a connection with the organisms I study,” she emphasizes – a factor that would be absent were she to work exclusively with abstract gene sequences.
Moreover, interpreting eDNA metabarcoding results is often not en-tirely straightforward. So Laakmann controls all species lists to check that the results make sense. If, for example, a species of zooplankton appears unexpectedly on a list, the researchers might need to check how trustworthy the database entries are. “In one case we worked out that a strange pattern in the data had been caused by a faulty identification using conventional methods,” she explains.
Why water samples contain DNA of terrestrial animals
The fact that land animals like chickens, cows, wild boar or mice keep turning up in the species lists is more of a curiosity. “Sometimes we can even work out why,” Laakmann adds. Genetic material from farmed animals like chickens and cows probably gets into the rivers, as do deposits from wild boar living in the estuaries, she explains. Ballast water is another potential source of exotic DNA: “But the mouse remains a mystery,” chuckles the biologist, who nonetheless intends to get to the bottom of the matter with Kingsly Chuo Beng.
Now, for the first time, we can identify which larvae are floating in the water at what time of the year
Silke Laakmann
Every now and then the researchers come across what are known as cryptic species – species that are genetically new but externally resemble other, already known species. Moreover, the genetic method increases the likelihood of finding rare or endangered species, including marine mammals. “We find more species than we could previously identify,” Laakmann stresses. All of these factors show the huge potential of eDNA in opening up previously hidden areas of biodiversity.
Procedure enables more precise determination
When it comes to the larvae found in the zooplankton samples of ben-thic organisms and fish, a whole new world is revealed. With conventional methods these miniscule organisms can often only be roughly assigned to the different animal groups.
Metabarcoding, however, allows research-ers to pinpoint what species many of these larvae belong to – provided they are already inventoried in a database. “Now, for the first time, we can identify which larvae float in the water at what time of the year. This allows us to draw conclusions about their distribution and reproductive cycles,” Laakmann comments. Particularly in late spring and summer, when many organisms reproduce, eDNA analysis can detect up to four times as many species in zooplankton samples than conventional methods.
The larvae of the European oyster are also almost impossible to identify under the microscope. At the free-swimming stage, when they are less than a millimetre in size, they resemble round blobs that move using lobe-like extensions, much like all other bivalve larvae. Laakmann and her team are nevertheless confident that they will be able to uncover the drift of the mollusc offspring in the vast expanses of the North Sea. In preparatory studies for the CREATE project they have already successfully identified the genetic material of the oyster larvae in water samples.
This story has first been published in the 2022/23 issue of the research magazine EINBLICKE.