Artificial intelligence could accelerate the search for an effective drug against Sars-CoV-2, says AI expert Oliver Kramer. He and his team have developed a method for identifying promising candidates.
Voice recognition, image analysis or online shopping: artificial intelligence (AI) already features in many products we use on a daily basis. Experts in this future-oriented technology also see great potential for its use in medicine. Image recognition software, for instance, could assist doctors in the diagnosis of cancer or bone fractures.
“But AI methods could also help to speed up the search for an effective active substance against the novel coronavirus Sars-CoV-2,“ says Prof. Dr. Oliver Kramer. A professor of computer science, Kramer leads the Computational Intelligence research group at the University of Oldenburg and studies AI techniques such as deep learning and evolutionary algorithms.
The targeted design of active substances, in other words the creation of chemical compounds for a specific purpose, was not the main focus of his work in the past. But when the coronavirus epidemic began to spread at the start of the year, Kramer decided to shift his attention to this field – also in order to show that in addition to its often controversial applications, AI research can contribute to resolving urgent problems in society.
Searching for active compounds
The search for active substances that can effectively combat a disease is currently a highly complex process that begins with laboratory tests and ends with clinical studies. The process generally takes several years to complete. To find suitable substances, pharmacologists first of all test the biological effects of thousands of potential active substances using fully automated procedures. In parallel, computer programmes are used to design substances with the most suitable physical properties, such as being able to block a specific biochemical reaction in a cell. AI methods could speed up this search, for example using self-learning algorithms that deliver ever better results over time. These methods are still in their infancy, however.
Together with five colleagues, Kramer has now developed an AI method for tracking down substances that can block a specific enzyme in the novel coronavirus Sars-CoV-2. However, developing a potential drug against Covid-19 is not the computer scientists’ primary objective. “Our contribution is above all a methodical approach in which we can apply our AI competences,” stresses Kramer.
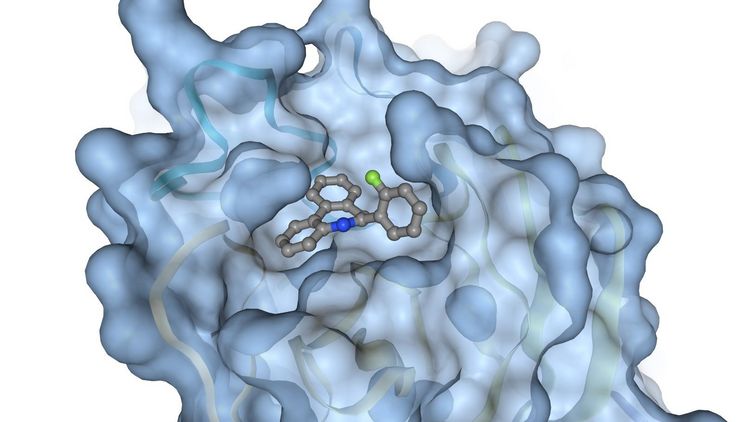
Modelling desired functions
The team, of which Thomas Teusch of the Theoretical Chemistry research group is also a member, chose the virus’s main protease as its target. The virus needs this enzyme as a replication tool: the protease is a large and complex biomolecule that breaks down long protein chains into smaller sections. Chinese researchers decoded the genome and the genetic blueprint of Sars-CoV-2 in January, and shortly afterwards a team of researchers at the University of Lübeck succeeded in elucidating the protease’s crystalline structure, thus fulfilling the prerequisite for developing an effective drug against the virus. “The idea is to find a molecule that binds to a specific site on the coronavirus protease and thus disrupts its function,” Kramer explains. Such a drug could interrupt the virus’s life cycle and thus ensure that the symptoms of the disease remain mild.
As is common in computer-based searches for active substances, the computer scientists represent chemical compounds in their model using character codes that precisely describe each individual atom and its position in the molecule. Their programme then tests the suitability of different substances on the basis of specific criteria. The researchers use an evolutionary procedure in their tests: beginning with ten to twenty starting substances, they create new compounds through random changes in the character code, which the programme then also tests for their suitability as an active substance. Out of each generation of compounds thus produced, the programme selects those with the properties best suited to creating the next generation of substances. The researchers let the programme run through this procedure 200 times on different settings, after which they analysed to what extent the resulting substances matched the desired criteria.
On the right path
“Our method is one of only a handful that optimise several properties of molecules at the same time, rather than just a single property,” explains Kramer. The candidate substances resulting from this procedure displayed certain typical properties of medications: many contained benzene rings, i.e. structures consisting of six carbon atoms, and most of the molecules were rather small, which is not untypical for effective protease inhibitors. Some substances that were able to bind to the Sars-CoV-2 protease particularly well had an unusual geometry.
These insights could at least be the starting point for further experiments, says Kramer. In his opinion, among other things the physical model for calculating how firmly potential protease inhibitors bind to the protease also needs to be improved in order to achieve more realistic results. For this purpose, the researchers used a standard programme that contains many simplifications in order to reduce the computing time.
When it came to publishing their results Kramer and his colleagues chose an increasingly popular option: they presented their study on a so-called preprint server and asked various experts for feedback. At the same time, they registered the article for presentation at an important conference. “It’s important to us that the method doesn’t disappear into a drawer. We want to make our work accessible to the scientific community, receive new inputs and then continue our research,” Kramer stresses. This is why the team decided to make its work public as quickly as possible. So far the feedback has been extremely positive: “The points of criticism that were brought to our attention pertained to minor details,” Kramer says. “This gives us hope that we’re on the right path.”