Research & Clinical Collaboration
Research & Clinical Collaboration
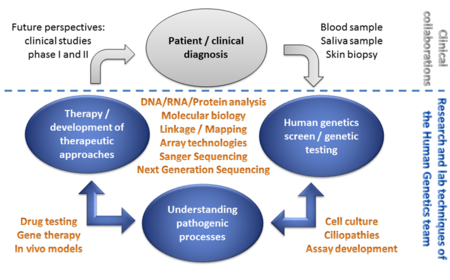
Aims and techniques
The ultimate goal is to help patients and families to better understand their disease and find effective treatment options.
We aim to understand the pathogenic mechanism underlying human genetic diseases and to develop therapies to treat the patient´s condition. We make use of patient material (e.g. blood samples and skin biopsies).
A multitude of technologies is applied to help families to understand the molecular basis of their diseases. Among others, these technologies include high-throughput sequence analysis (Next Generation Sequencing) or Sanger Sequencing, and different technologies originating from molecular biology, cell culture and therapy development. A detailed characterization of the pathogenic processes associated with genetic alterations is performed by detailed transcript analyses, in cell culture assays, or in in vivo disease models. The results of these analyses build the basis to develop therapeutic approaches.
Human Genetics
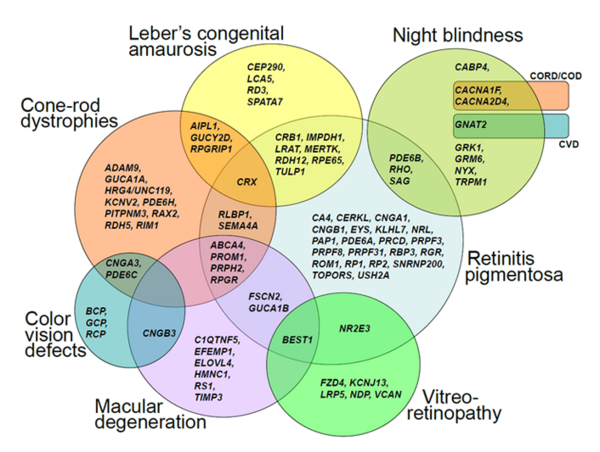
We are broadly interested in the genetic and molecular basis of human diseases. Our research focus includes ciliopathies, retinal dystrophies, hearing deficits and brain malformations, all of which are among the most heterogeneous genetic diseases known so far. We analyze family pedigrees and search for genetic causes of inherited disorders.
Genetic heterogeneity and clinical variability are obstacles to the identification of disease-associated genes. Next Generation Sequencing is the method of choice to overcome these obstacles. It enables the analysis of all known genes (whole exome or genome sequencing) in parallel.
Pathomechanism
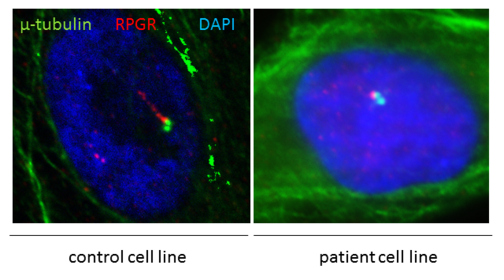
We aim to understand the pathogenic processes underlying genetic diseases. After identifying pathogenic sequence variants (mutations) in disease-associated or novel candidate genes, the biological function of the disease genes is characterized. We culture patient-derived cell lines or make use of in vivo disease models to identify disease-relevant mechanisms. Additional examples of techniques applied to study the consequence of mutations include DNA/RNA/protein analyses, different cell culture techniques, and immunohistochemical stainings.
Frequently, mutations in disease associated genes belong to the group of ciliopathies. RPGR mutations cause ciliopathies (see figure) and are associated with retinal degenerations in patients.
Therapy Development
Research labs around the world develop therapeutic approaches to treat genetic diseases. Replacing the defective gene by a functional copy (replacement therapy) is one of the more common therapeutic strategies. Furthermore, we successfully applied therapeutic strategies to treat the underlying genetic defects and corrected the ciliopathy phenotype in patient-derived cell lines and in vivo models.
We develop gene therapeutic approaches in the lab. One example is described below and uses techniques to correct defective transcript processing:
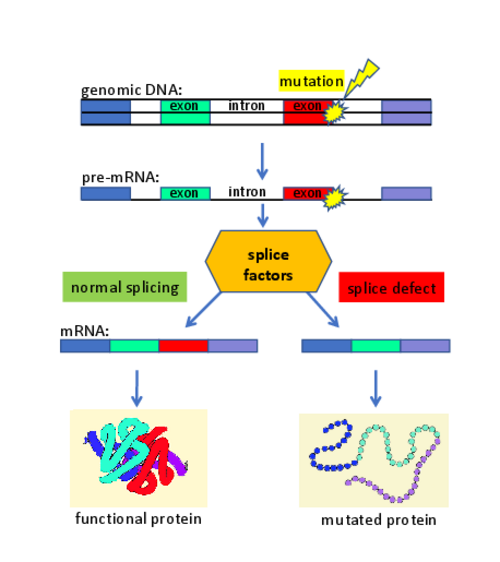
On average, about 20% of the patients show disease-causing mutations that lead to defects in the splicing of genes. This observation is largely independent from the affected disease gene. Thus, the pathogenic effect of many mutations in monogenetic diseases primarily alters the gene transcript (and only subsequently affects the protein composition). We developed gene therapies to treat diseases that are caused by mutations affecting the splice process. We used splice factors (e.g. U-snRNAs) and designed them to correct the patient-specific splice defects. Splicing assays (using antisense-oligonucleotides (AONs), minigenes, or patient-derived cell lines) were utilized to optimize the gene therapeutic approaches. We demonstrated that these therapeutic strategies are functional in patient-derived cell lines and in vivo models of diseases.
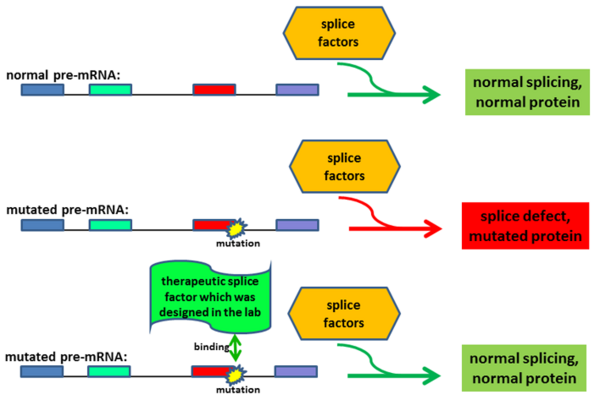
Research labs around the world develop therapeutic approaches to treat genetic diseases. Replacing the defective gene by a functional copy (replacement therapy) is one of the more common therapeutic strategies. Furthermore, we successfully applied therapeutic strategies to treat the underlying genetic defects and corrected the ciliopathy phenotype in patient-derived cell lines and in vivo models.
We develop gene therapeutic approaches in the lab. One example is described below and uses techniques to correct defective transcript processing:

On average, about 20% of the patients show disease-causing mutations that lead to defects in the splicing of genes. This observation is largely independent from the affected disease gene. Thus, the pathogenic effect of many mutations in monogenetic diseases primarily alters the gene transcript (and only subsequently affects the protein composition). We developed gene therapies to treat diseases that are caused by mutations affecting the splice process. We used splice factors (e.g. U-snRNAs) and designed them to correct the patient-specific splice defects. Splicing assays (using antisense-oligonucleotides (AONs), minigenes, or patient-derived cell lines) were utilized to optimize the gene therapeutic approaches. We demonstrated that these therapeutic strategies are functional in patient-derived cell lines and in vivo models of diseases.


