Contact
Supertrees and phyloinformatics
Supertrees and phyloinformatics
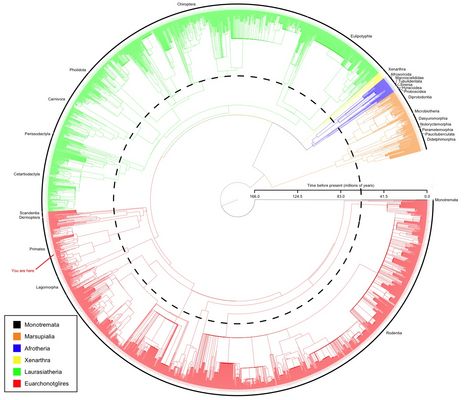
The molecular revolution combined with the availability of sequence data in public databases such as GenBank, Swiss-Prot, or HOVERGEN has increased the scope of phylogenetic inference tremendously. Never before has so much informative data been available. However, the wave of new data has also brought a wave of new questions and problems: how can we best analyze the data so as to achieve the most comprehensive and robust phylogenies possible?
Supertree construction
|
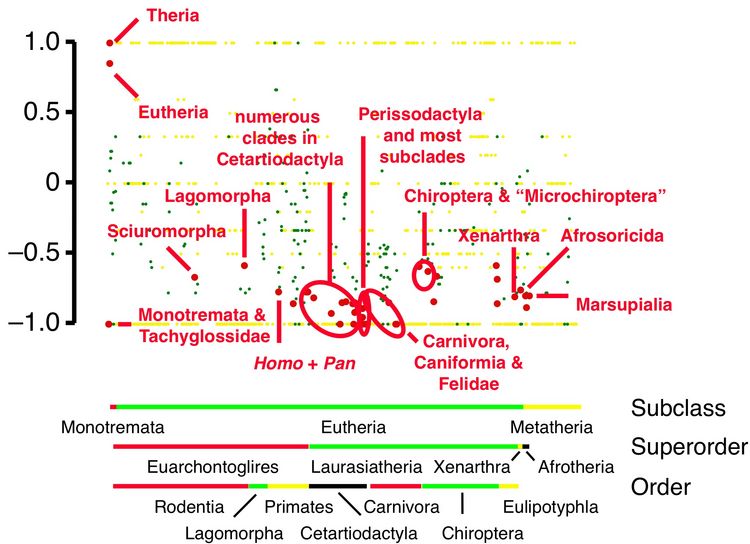
Phyloinformatics
|