Research
Ecology of microorganisms in Wadden Sea and beach sediments

Marine sediments in the coastal areas of the
North Sea are characterized by high productivity,
strong chemical gradients and a high microbial
diversity. Our goal is to record this diversity
and to assess the ecological importance of the
different microorganisms.
Cultivation of yet uncultivated marine microorganisms
Only 1 % of the existing microorganisms could be isolated in pure cultures so far. In order to expand our knowledge of the physiology, biochemistry and ecology of previously uncultivated groups of organisms, we are developing novel cultivation concepts.
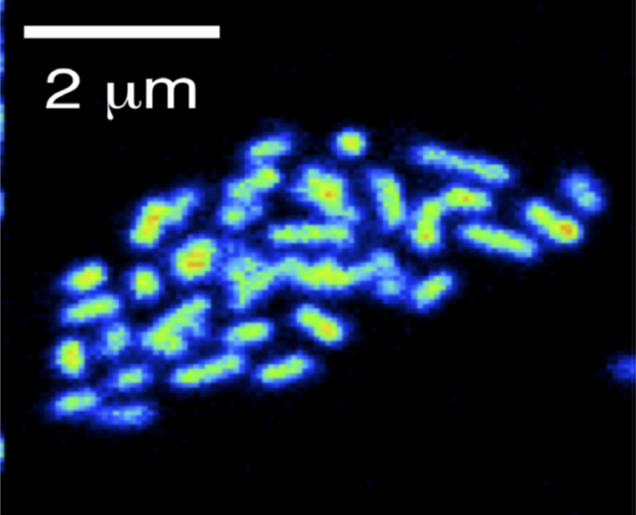
Physiology and biogeochemical importance of Archaea in the oceans

Microorganisms of the domain Archaea have long been considered extremophiles, found only in a few habitats. In fact, archaea colonize many marine habitats. In our group we investigate the physiological properties of marine archaea and determine their metabolic performance within biogeochemical processes.
Microbiology of the marine deep biosphere
The deep regions of marine sediments form one of the largest habitats for microorganisms. One of the goals of our research is to investigate microbial survival strategies in these difficult-to-access habitats.


![[Translate to English:]](/f/5/_processed_/3/2/csm_ICBM-Logo-transparent-_91fe1c6774.png)