Contact
Dr. Bert Engelen

Institute for Chemistry and Biology of the Marine Environment (» Postal address)
Carl von Ossietzky Universität Oldenburg
Ammerländer Heerstraße 114-118
26129 Oldenburg
W15 1-122, Carl-von-Ossietzky-Str. 9 - 11 (» Adress and map)
Fr 10 - 12 Uhr
Prof. Dr. Heribert Cypionka
Email: Cypionka[at]icbm.de
Other projects (1997-2018)
Achromatium
Achromatium is a genus of unicellular sulfur bacteria, which appears in freshwater and brackish water sediments all over the world. With a length of 15 µm to more than 100 µm and a width of up to 40 µm (Head et al. 1996) it belongs to the giant bacteria, like Beggiatoa or Thiomargarita namibiensis. In 1903, the first species of Achromatium was described. Until now it is not possible to culture these bacteria in the laboratory.

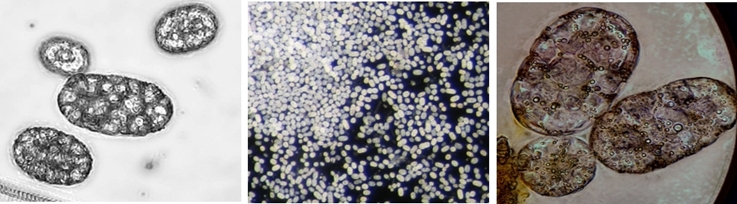
Left: Achromatium cells collected manually by pipette in a small bowl (4 cm) from a water sample. Right: Manually collected Achromatium cells (40x)
The bacterium gains energy from oxidising sulfide to sulfate. In addition to that, it fixes inorganic carbon. Because of this metabolic pathway, the intracellular structure of Achromatium oxaliferum consists of elemental sulfur droplets. Another characteristic is the occurrence of large calcium carbonate crystals within the cell. Their function is under discussion till now. Maybe the carbonate crystals maximise the ratio of the cell surface to the volume of the cytoplasma, so the substrate uptake and the transport limitation can be reduced (Head et al. 2000). Another possibility is that they are responsible for the buffering of pH or to support a high pressure of CO2 to enable the inorganic carbon fixation (Head et al. 1996).
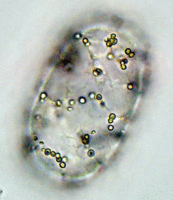
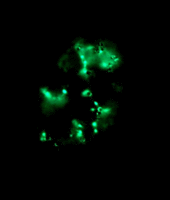
Left: Image stack of Achromatium cell in light microscopy. Right: Image stack of the same cell in fluorescence microscopy, stained with Sybr Green I.
By staining with DAPI or Sybr Green, epibionts on the surface of the Achromatium cell can be visualized. Also the DNA inside the cells can be shown. It is conspicuous, that the DNA is not distributed consistent in the whole cell, but concentrated in one part of it.
Current Objectives
Although the genus Achromatium was discovered more than one century ago, there are still a lot of questions to be answered. Our current work focuses on the DNA distribution inside the cell. First visualizations with Sybr Green I suggest specific regions and not a random distribution of DNA inside the cell. Furthermore, we try get an overview about the epibionts, before and after washing, by sequencing. This is a necessary step on the way to completely sequence achromatium.
Team
- Heribert Cypionka
- Hans-Peter Grossart
- Sina Schorn
Related literature
- Head, I.M., Gray, N.D., Clarke, K.J., Pickup, R.W., Jones, J.G., The phylogenetic po-sition and ultrastructure of the uncultured Bacterium Achromatium oxaliferum. Micro-biology (1996) 142, 2341-2354
- Head, IM., Gray, ND., Babenzien, H-D., Glöckner, FO., Uncultured giant sulfur bacteria of the genus Achromatium. FEMS Microbiology Ecology 33 (2000) 171-180
Links
- Mikrobiologischer Garten
Ecophysiology of ammonia-oxidizing Archaea
Background
The isolation of a marine mesophilic crenarchaeote that grows chemolithoautotrophically by aerobically oxidizing ammonia to nitrite (Könneke et al. 2005) was lately described. The strain, named candidatus Nitrosopumilus maritimus, represents the first nitrifier within the domain Archaea. This study also correlated for the first time the presence of archaeal ammonia-monooxygenase (AMO)-encoding genes with the biological conversion of ammonia to nitrite. The archaeal amoA gene was subsequently used as molecular target to demonstrate the ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean (Francis et al. 2005). Analysis of this gene also indicated that archaeal ammonia oxidizers are more abundant in soil than their well-known bacterial counterparts (Leininger et al. 2006). Similar findings were recently obtained from natural water samples of the North Sea and the Atlantic Ocean where archaeal amoA genes were detected in copy numbers up to three orders of magnitudes higher than those of bacterial amoA genes (Wuchter et al. 2006). Estimates based on biomass production confirmed the hypothesis that Crenarchaeota might be the most abundant nitrifiers in the ocean (Ingalls et al. 2006). Nitrification is a central biological process in the global nitrogen cycles. The complete oxidation of ammonia, has been thought so far, involves only two physiological distinct groups of microorganisms, the ammonia-oxidizing bacteria (AOB) and the nitrite-oxidizing bacteria (NOB) (Bock and Wagner, 2000. All cultured nitrifiers are chemolithoautotrophs and obligate aerobes. Most of the AOB and NOB fix inorganic carbon by the Calvin-Benson pathway. Only a few fix CO2 by the phosphoenolpyruvate carboxylase (Harms et al. 1981, Takahashi et al 1993). Based on tracer studies it is suggested that the oxidation of ammonia to nitrite is the rate-limiting step of nitrification in nature (Bock and Wagner, 2000). The discovery of archaeal ammonia-oxidation in N. maritimus might now challenge the current perspective on biological nitrification and the impact of AOB on the nitrogen cycle (Nicol and Schleper, 2006). Nitrogen is a limiting factor in many ecosystems, and the abundant crenarchaeal nitrifiers represent key players in the competition for ammonia or other reduced nitrogen compounds. Furthermore, The global crenarchaeal inorganic carbon fixation in the ocean was estimated between 3.3 and 6.5 · 1013 mol carbon per year, accounting for approximately one percent of the annual marine primary production (Herndl et al. 2005, Ingalls et al. 2006, Wuchter et al. 2006).
Goals
The main goal of our research is to understand the ecophysiology of ammonia-oxidizing archaea. We aim to discover biochemical pathways involved in the chemolithoautotrophic lifestyle of the only existing pure culture. The studies are being performed in close collaboration with David Stahl and colleagues at the University of Washington and are facilitated by the ongoing whole genome analysis at the Joint Genome Institute. Biochemical and classical microbiological analysis are being applied to investigate the mechanisms mechanisms of carbon metabolism and to approve hypothetical pathways.
The phylogenetic diversity of archaeal amoA and 16S rRNA-genes suggests the existence of various physiotypes. A current project in our lab deals with the isolation of further AOA strains from aquatic and terrestrial environments where the corresponding biomarkers have prevoiusly been detected. The new strains will be characterized physiologically and will help to understand the suggested high impact on the biogeochemical nitrogen and carbon cycles.
Team
- Martin Könneke
- Sonja Standfest
- Heribert Cypionka
Collaboration
- University of Washington, Seattle, USA (David A. Stahl, José de la Torre, Willm Martens Habbena, Chris Walker)
- IFM-GEOMAR, Kiel (Michael Hügler)
- WHOI, Woods Hole, USA (Stefan Sievert)
- NIOZ, Den Burg, The Netherlands (Jaap Sinninghe Damsté, Stefan Schouten)
- Radboud University, Nijmegen, The Netherlands (Huub Obdencamp, Suzanne Haaijer)
Related publications
- De la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl D (2008) Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10:810–818
- Könneke M (2008) New Crenarchaeota - the organismic link in the nitrogen cycle. In: Amann R, Goebel W, Reinhold-Hurek B, Schink B, Widdel F; (Eds.) Life strategies of microorganisms in the environment and in host organisms, Nova Acta Leopoldina NF 96, Nr. 356, ISBN: 978--3-8047-2499-0, Suppl. 3-8
- Schouten S, Hopmans EC, Baas M, Boumann H, Standfest S, Könneke M, Stahl DA, Damste JSS (2008) Intact membrane lipids of "Candidatus Nitrosopumilus maritimus", a cultivated representative of the cosmopolitan mesophilic group I Crenarchaeota. Appl Environ Microbiol 74:2433-2440
- Könneke M., Bernhard, A. E., de la Torré, J. R., Walker, C. B., Waterbury, J. B., Stahl, D. A. (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543-546
- Martin Könneke (Oral presentation, Leopoldina Symposium: Life strategies of microorganisms in the environment and in host organisms, 2006) New Crenarchaeota - The organismic link in the nitrogen cycle
- Sonja Standfest, Heribert Cypionka and Martin Könneke (Poster, VAAM Annual-meeting 2007) Determination of growth characteristics of the ammonia-oxidizing crenarchaeon candidatus "Nitrosopumilus maritimus"
- Martin Könneke (Oral presentation, ASM General meeting 2007) Nitrifying archaea.
- Ellen C. HOPMANS, Stefan SCHOUTEN, Marianne BAAS, Henry BOUMANN, Sonja STANDFEST, Martin Könneke, David A. STAHL, and Jaap S. SINNINGHE DAMSTÉ (Poster, IMOG 2007)Membrane lipids of Candidatus "Nitrosopumilus maritimus", a cultivated isolate from the ubiquitous marine Crenarchaeota
Methane release from coal seams
Background
About seven percent of the global annual methane emissions originate from coal mining. Furthermore, during the last decade the use of coal bed methane has come into focus of the power producing industry. In Germany, the gas from active and abandoned mining areas is increasingly used for heat and power production, especially after the introduction of the "renewable energy law" in 2000. In many coal reservoirs worldwide, the analysis of the stable carbon isotopic composition of methane showed that the produced methane is a mixture of thermogenic and biogenic origin. However, the timepoint for the formation of the latter fraction is not known. Interestingly, time series of measurements show an increasing proportion of the biogenic, microbially produced methane during the last years, indicating a recent origin.
First experiments
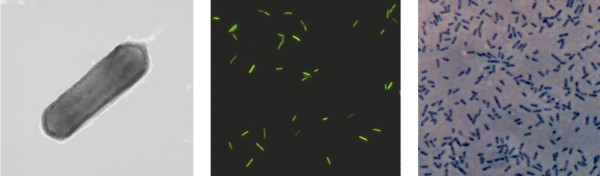
Incubations of wood and coal samples from abandoned reservoirs showed over a period of nine months constant and significant microbial methane production rates. The stable carbon isotope signatures of the produced methane were in a similar negative range as the values of the gas samples collected in situ in the reservoir. Fluorescence-in-situ-hybridisation (FISH) revealed the presence of different types of methanogenic Archaea. These could be further enriched with common methanogenic substrates, like acetate, hydrogen/carbon dioxide or methanol. Detailed molecular studies are currently carried out to identify the microorganisms involved in this scientifically as well as economically very important process.
Team
- Sabrina Beckmann
- Bert Engelen
- Heribert Cypionka
Cooperation
- Martin Krüger (BGR)
Related publications
- Beckmann, Sabrina (2011) Dissertation: Microbial Methane Formation in Abandoned Coal Mines in the Ruhr Basin of Germany
- Beckmann S, Lüders T, Krüger M, von Netzer F, Engelen B, Cypionka H (2011) Acetogens and acetoclastic methanosarcinales govern methane formation in abandoned coal mines. Appl. Environ. Microbiol. 77:3749–3756
- Beckmann S, Krüger M, Engelen B, Gorbushina AA, Cypionka H (2011) Role of Bacteria, Archaea and Fungi involved in methane release in abandoned coal mines. Geomcrobiol J 28:347-358
- Krüger M, Beckmann S, Engelen B, Thielemann T, Cramer B, Schippers A, and Cypionka H (2008) Microbial methane formation from hard coal and timber in an abandoned coal mine. Geomicrobiology J. 25:315-321
- Thielemann T, Cramer B, and Schippers A (2004) Coalbed methane in the Ruhr Basin, Germany: a renewable energy resource? Organic Geochemistry 35, 1537-1549
Links
- Pictures from the 2007 sampling campaign
Aerotaxis in Desulfovibrio
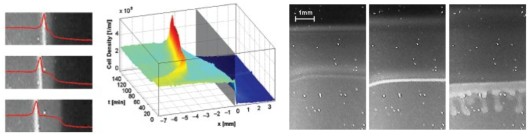
Aerotactic band formation by Desulfovibrio desulfuricans (DSM 9104) was studied in a stopped-flow diffusion chamber. This chamber allowed us to create reproducible, steep oxygen gradients in a flat capillary, time-lapse video recordings and spatio-temporal analysis of band formation. The cells formed two types of bands. Bands of the first type evolved quickly after starting the experiment and were located near the oxic–anoxic interface. Bands of the second type typically appeared several minutes later and a few millimeters inside the initially anoxic volume of the capillary. Band formation depended on metabolism and could be stimulated by lactate addition, and thus appears to be energy taxis. Mathematical modeling of oxygen diffusion and respiration within the chamber revealed that bands formed preferentially at oxygen concentrations close to 4% air saturation. The swimming speed of the cells was determined by digital single-cell tracking and found to be highest close to the oxic–anoxic interfaces. Motility patterns were influenced by surfaces, at which cells accumulated. Bioconvection sometimes occurred if very dense bands had formed. The ecological implications of these two phenomena are unknown.
Team
- Jan Fischer
- Andrea Eschemann
- Andrea Sass
- Henrik Sass
- Heribert Cypionka
Cooperations
- Michael Kühl
- Roland Thar
Related publications
- Fischer J, Cypionka H (2006) Analysis of aerotactic band formation by Desulfovibrio desulfuricans in a stopped-flow diffusion chamber. FEMS Microbiol Ecol 55:186-194
- Sass AM, Eschemann A, Kühl M, Thar R, Sass H, Cypionka H (2002) Growth and chemosensory behavior of sulfate-reducing bacteria in oxygen-sulfide gradients. FEMS Microbiol Ecol 40:47-54
- Eschemann, A, Kühl M, Cypionka H (1999) Aerotaxis in Desulfovibrio. Environ Microbiol 1:489-495
Links
- Microbiological garden
Nevskia ramosa
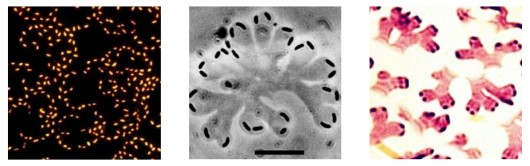
Seventy percent of our earth is covered by water. The water-air interface represents a special habitat, as it accumulates organic compounds and microorganisms due to the high surface tension of water. The microbial community at the water surface is named neuston. The most famous neuston bacterium is Nevskia ramosa. The bacteria form flat rosette-like structures that branch when the cells divide. On the water surface, the bacteria are exposed to high doses of UV light. However, they are resistant to those by special repair mechanisms of DNA damages. They are feeding on simple organic molecules as lactic acid or acetic acid. Their special purpose of life on the water surface appears to be that they can collect nitrogen compounds entering the water from the air. If nitrogen compounds are added to their growth medium, they dive down and grow within the water body.
Team
- Heike Stürmeyer, now Eilers
- Teresa Pladdies
- Jörg Overmann
- Heribert Cypionka
Cooperations
- Rudolf Amann, MPI Marine Mikrobiologie, Bremen
- Hans-Dietrich Babenzien, IGB Berlin/Neuglobsow
- Frank-Oliver Glöckner, MPI Marine Mikrobiologie, Bremen
Related publications
- Pladdies T, Babenzien H-D, Cypionka H (2004) Distribution of Nevskia ramosa and other rosette-forming neustonic bacteria. Microb Ecol 47:218-223
- Stürmeyer H, Overmann J, Babenzien HD, Cypionka H (1998) Ecophysiological and phylogenetic studies of Nevskia ramosa in pure culture. Appl Environ Microbiol 64:1890-1894
- Ottenjann T (1998) Vergleich von Rosetten-bildenden Bakterien asus dem Neuston verschiedener Gewässer. Diplom-Arbeit, Univ. Oldenburg
- Stürmeyer H (1997) Isolierung und mikrobiologische Charakterisierung von Nevskia ramosa. Diplomarbeit, Univ. Oldenburg
- Glöckner FO, Babenzien HD, Amann R (1998) Phylogeny and identification in situ of Nevskia ramosa. Appl Environ Microbiol 64:1895-1901
Links
- Nesvkia in the Microbiological Garden

![[Translate to English:]](/f/5/_processed_/3/2/csm_ICBM-Logo-transparent-_91fe1c6774.png)