Usermanual HTML
User Guidelines
(Beta-Version, 01.12.2019)
Checklist before starting:
- I have read the publication and user guidelines associated with ICBM-OCEAN.
- My spectra are sufficiently mass calibrated (mass error < 1 ppm) and in the requested format for use in ICBM-OCEAN. If I want to start MDL calculations, I have exported my data at a SNR of 0 from preceding software. Starting with Step 1 (Method detection limit) this means my data has the following format: m/z (mass), I (intensity), S/N (signal to noise based on preceding software), Res. Or ResPow (resolution power). For m/z I supply a precision of at least 5 decimals.
m/z I S/N Res. 92.1424641 854283 1.4 2713601 - I understand that ICBM-OCEAN is a free software that will undergo continuous further development. It comes without any warranty. If used for publication or else, I will acknowledge ICBM-OCEAN as stated in the site notice including the version number and the settings I used.
From the ICBM-OCEAN main page (www.icbm.de/ICBM-OCEAN) click on the start button to initiate a new data processing operation. You will be prompted with a dialogue of checkboxes inquiring some basic preconditions for the subsequent data handling (e.g. whether your data input meets the requirements for a good result).
Example data for all steps of the formula attribution process are available for download on the ICBM-OCEAN Start page.
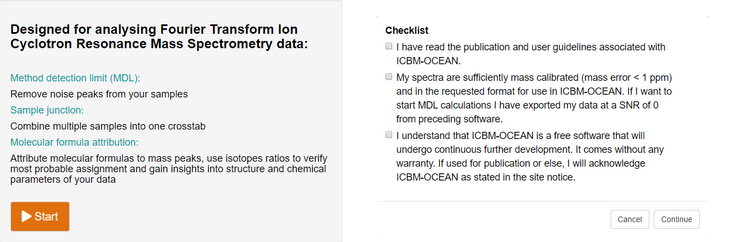
Figure 1: Before you start with your ICBM-OCEAN data processing you will be prompted to check some premises for the evaluation to guarantee that you will arrive at an optimal result but also acknowledge and are aware of the software’s limitations.
Note: ICBM-OCEAN is designed to be used directly in the browser (online). For very large datasets calculations can be done offline. For this you must upload your data as in the online mode but click the “Run offline” button in the MDL page (which is the first step, see next paragraph). For the offline mode you must supply your e-mail address as well as a project name and type (e.g. Marine DOM, Terrestrial DOM). Results will be calculated with the settings you chose in the user interface (on all pages (MDL, sample junction, formula attribution)!). Alternatively you can upload a profile that you created. For creating a profile select all your settings in all tabs, go to the “site notice” tab and scroll down to the “save profile” button. After your data is processed in the offline mode, you will receive an email with a link to download the respective results. The offline mode currently does only raw calculations but does not supply all steps e.g. interactive validation plots as in the online mode. We recommend using the online mode if your data is below 1 GB.
Step 1. Method detection limit (MDL)
1.1 Tab “MDL”:
Load your calibrated masslists into the tool by clicking the browse button. Make sure the “csv” files you want to upload are separated by the separator selected in the radio buttons below the browse field before clicking. Masslists should contain m/z (m/z), S/N (S/N), resolving power (ResPow) and intensity (I) with the column names given in the brackets. Select all the files you want to include in your analysis. The order in which they appear in the folder will be the order in which they will be processed. Noise estimation in this step can be done based on instrumental blanks, operational blanks or it is directly derived from the samples. If blank analyses are available, the respective files have to meet certain naming conventions to be recognized by the system, i.e. the substrings “_blank_instrumental” or “_blank_operational” must be included.
Click “open”. The files will be uploaded.
Figure 2: MDL calculation options in ICBM-OCEAN following file upload.
After the message “upload complete” appears below the browse button, new fields will show up. If you are an Orbitrap user, you can do a single mass shift by selecting the respective checkbox and filling in the mass shift parameters. If you are willing to supply your data for a later meta-analysis of datasets processed by ICBM-OCEAN, select the “Agree to archive for meta-analysis:” checkbox. This is not mandatory, but will allow us to save the dataset on our server. If the user agrees, he will be included in a meta-analysis publication when enough data is collected globally. Otherwise (if not agreed) supplied datasets will be deleted from the server after the browser is closed. The option to remove outliers of suspicious mass vs. resolution power pairs is able to eliminate outliers like side-peaks. It is based on the first derivative of a kernel density estimation applied on the residuals in a median regression.
Click “run”. MDL calculation will be performed.
When MDL calculation is completed, two plots will appear that show the result of the respective calculation (Figures 3 and 4) and help to find a suitable MDL level for your data. The blue lines should show a monotonic increase. If the fitted detection limit seems too high or shows a strong curvature, you may select the number of samples with the highest noise from the table to be excluded from the analysis and click run again.
Figure 3: Output of an MDL calculation based on included operational blanks. Displayed are the noise peaks (black dots) and the derived MDL levels that will be used to distinguish between true peaks and noise.
By clicking the respective download buttons, you can choose which results you want to download (e.g. MDL1, MDL2, MDL3). Which one you choose depends on your preference of minimizing type I or type II error: At higher MDL level, the risk to include false positives decreases (Figure 4c), while at the same time the risk to exclude true peaks as noise increases.
Figure 4: (a) Q-Q plot of the noise (green) and sample quantiles (yellow). The point where these two diverge should be considered as the minimum MDL to be chosen for further processing. (b) Estimated ratio of noise to total peaks. (c) Approximation of false positives to expect for different MDL levels. Panels d-f show results of outlier removal. Panel g shows how side peaks flank a true peak. Such side peaks can be eliminated by ICBM-OCEAN during processing (d-f).
1.2 Tab “Deselect contaminations”:
After MDL calculation took place a plot will appear inside this tab (Figure 5). Notice: the plot might take some time to load, depending on the size of your data. By brushing points inside the plot and clicking on the “toggle points” button, peaks can be excluded for further analysis.
Figure 5: Manual deselection of contaminants after MDL calculation.
1.3 Recommended settings
For our example we recommend using MDL2.5 (see text Figure 4).
Step 2. Sample junction
2.1 Tab “Sample junction”
Either proceed with your MDL data, selecting one of the options in the radio buttons or load a new datafile into the tool by clicking the browse button and “File Input”. Choose the maximum tolerance at which masses from different samples should be merged. Select your joining method, either “fast join” or “precise join”; note that precise join will be much slower. If you want to recalibrate your masses, click on the “Use recalibration” checkbox and select the elements and their respective ranges and a maximum tolerance for a formula attribution needed inside the recalibration process as well as other settings e.g. the analyzing mode (positive, negative). Select the recalibration method you want to use as well as whether you want to do a median or mean fit for recalibration along the mass axis. The results of a recalibration or sample junction can be evaluated from error visualizations (Figure 7 and 8). Default tolerances for recalibration and junction are both 0.5 ppm.
Figure 6: Sample junction options in ICBM-OCEAN following input of MDL data.
The results can be downloaded with the “Download” button.
Figure 7: Optional recalibration to correct for systematic error in mass accuracy. a) If mass error is plotted against m/z, a significant systematic error will manifest as a non-linear trend (original sample). b) The recalibration will minimize the error by reconstructing the systematic bias along the mass axis and subtracting it for each mass (adjusted sample).
2.2 Recommended profile of settings
Use “standard smooth” if you are an FT-ICR-MS user. If you are an Orbitrap user and smoothing results are not sufficient, try “tensor product smooth” and run again. Although this method is currently in a beta stage it might optimize results for Orbitrap users.
If your data shows increasing variance along the mass axis, you can select “use median” or/and use “log diff” to cope with that.
We recommend using “fast join” whenever possible.
Figure 8: Sample junction mass merging with different tolerances. a) If tolerance is chosen too high the junction will result in a bimodal distribution. Here, slightly deviating masses in different samples (due to instrumental variability) which actually correspond to identical molecular formulae are not properly merged. B) If tolerance is chosen well, the result will show a unimodal distribution. c) with a well-chosen tolerance the nearest neighbor distance distribution will at least be bimodal and Hartigan’s Dip test will be significant.
Step 3. Molecular formula attribution
3.1 Tab “Formula attribution”
You can either proceed from sample junction or load a new file (sample junction output). Use the respective radio buttons to select which data you want to use. Select elements and ranges that you want to use for molecular formula attribution. The tab “more” includes additional elements.
Figure 9: Formula attribution options in ICBM-OCEAN following input of sample junction data.
Select the tolerance you want to allow for formula attribution, as well as mode (positive, negative) and the filters you want to apply to find the likeliest molecular formula (see Table 1; for details see ICBM-OCEAN publication). The tab “Contaminations” allows uploading a list of known contaminants that are deleted from the dataset at the end of the formula attribution process. Click on run. Do not click the checkbox “Reinsert 13C & 15N isotopes. Use only in isotope enrichment experiments. Caution: can produce wrong results“, unless you are planning to do enrichment experiments, in communication with the Marine Geochemistry group.
A result table will appear listing all attributed formulae as well as Van Krevelen diagrams (Figures 10 and 11) and a table with a statistical summary how many molecular formulae could be attributed or are isotope verified relative to the likeliest matches. The mass displayed in the table refers to the neutral molecule, not the measured ionized compound. If the homologous series filter option is used, the respective network can be visualized by selecting a formula from the table and clicking “show network” (Figure 12). In the next plot, the isotope ratio deviance for the element selected in the box below the plot (Figure 13) is visualized in a violin plot. Inside the select box “Choose result:” you can switch between different results, influencing the table and Van Krevelen diagram (see Table 2). Depending on what is selected here, the download button on the lower left of the page can be used to download the chosen results.
Figure 10: Van Krevelen diagram showing all isotope verified likeliest matches in an example dataset while color coding by individual aromaticity indices (AImod). The number of depicted formulae can be confined by certain criteria (aromatic, highly unsaturated, unsaturated, saturated compounds; oxygen-poor or -rich). Other characteristics can be color coded, i.e. double bond equivalents (DBE) or aromaticity (AI). For details on the definition of the different groups and characteristics see ICBM-OCEAN publication.
Figure 11: Frequency distribution of all isotope verified likeliest matches in van Krevelen space.
Figure 12: Homologous series network for C12H10O2 considering CH2 and O structural components.
Figure 13: Isotope ratio deviance of 13C before (right) and after (left) filtering and processing for likeliest match data.
3.2 Tab “Crosstab processing”
Here you can find some descriptive plots (Figure 14), e.g. a plot of matched masses against intensity, the summed intensities per sample, results from the linear quantile regression and the error partitioning filter.
Figure 14: Descriptive plots that allow for an in-depth evaluation of the different processing operations applied to the dataset. Panels a and b depict the distribution of all likeliest matches as well as the respective absolute mass error vs. S/MDL. Ideally the error should decrease with increasing S/MDL. Panel c shows the result of the linear quantile regression filter, all masses outside the set range were excluded from the dataset. d) shows the summed intensities per sample (after MDL).
Table 1: Explanation of parameters for formula attribution.
| Parameter | Calculation |
| N, S, P rule | This filter removes “unlikely” molecular formula assignments containing more than 3 heteroelements, excluding N3 and N4. |
| Delete Singlets | This filter will remove all masses that only occur in a single measurement prior to formula assignments (e.g. for processing datasets with replicated analyzes; recommended for datasets with > 5 spectra) |
| Linear quantile regression | This filter optimizes formula assignment by using statistical assumptions. The systematic mass error, i.e. the deviation between measured value and theoretical value can be assumed to be normally distributed. With linear quantile regression, an upper and lower percentile (Q1: 0.25 and Q3: 0.75) are defined. The 25 % of formulae corresponding to the masses deviating the most from the theoretical value above and below these thresholds (defined by the inter quartile range) are dismissed as unlikely. |
| Error partitioning filter | This filter optimizes formula assignment by using statistical assumptions. A set of peaks with unequivocally assigned formulae and thus known reference masses is used to compute a function that describes the mass error of the measurement. The error can constitute confidence intervals around a reference mass that should contain 95% of slightly deviating masses (Gaussian distribution) corresponding to one formula. Likewise, formulae assigned to masses outside the confidence range can be excluded. |
| U. oxidation state of metals | This filter eliminates molecular formulae containing metals with different oxidation states e.g. Cu_I and Cu_II |
| Isotope verification (13C (1-2), 15N, 18O, 34S, 81Br, 37Cl, 54Fe, 65Cu, 66Zn, 60Ni) | The filter makes use of natural isotope abundances (e.g. 12C vs. 13C) to optimize formulae attribution. Intensity ratios of two masses corresponding to two isotopologues (i.e. identical formulae but at least one atom has a different number of neutrans ) should reflect natural isotope abundances, i.e. a certain δ-value should be attached. This option checks for plausibility of this ratio for a mass pair while allowing for an adjustable δ-value tolerance and an eventual isotope verification of the respective formulae. In order to keep low intensity peaks – for which the chance of false elimination by this method increases – an S/MDL limit can be adjusted. By checking the “use mean” checkbox the average isotope ratio for each isotopologue mass pair across all samples is used, otherwise each sample is tested separately. For C, the option “second 13C” allows for an advanced isotope verification that includes a potential 3rd mass, corresponding to an isotopologue in which two 12C atoms are substituted by 13C isotopes. |
| Homologous series | If a given mass can be assigned to multiple formulae, the most likely one is determined to be the one with the largest corresponding homologues series network, i.e. formula groups that only differ by multiples of a structural component (i.e. CH2, CO2, O, H2, H2O). |
Table 2: Explanation of possible data downloads.
| Result | Contains |
| All matches in interval | All matches surviving the isotope ratio filters & error partitioning (but still including multiple chemical formulae for a single mass) |
| Likeliest match (isotope) | The likeliest chemical formula for a given mass based on our filters applied. A likeliest formula is chosen by the following ranking:
This selector can be further specified to filter certain subgroups, i.e. (1) formulae that only contain C and H, (2) CHO compounds without hetero atoms or (3 - 5) only formulae that also contain N, S or P. |
| Isotope verified (likeliest match) | Subset of the likeliest match data including isotope verified chemical formulae only. This selector can be further specified to filter certain subgroups, i.e. (1) formulae that only contain C and H, (2) CHO compounds without hetero atoms or (3 - 5) only formulae that also contain N, S or P |
| Isotope verified plus network | Subset of the likeliest match data including isotope verified chemical formulae and their homologous series network partners. |
| No matches | List of masses to which no molecular formula within the specified tolerances and elemental compositions could be assigned |
| Suffix “all under threshold) | These subsets are additional restrictions to the above explained subsets. These are only showing isotope verified formulae that are always below the chosen threshold. Exemplarily, a formula with δ13C of -30 and δ15N of -100 would be isotope verified for δ13C when the tolerance is 50 permille. So it will appear in the classic isotope verified subset but not in the subset isotope verified (all under threshold) that requires also δ15N to be inside the set tolerance ranges of 50 permille. |
3.3 Tab “Scaling and Ordination”
This tab should give a first insight into similarities between samples and which molecular formulae determine the major differences.
Select the type of linkage method you want to use for a hierarchical clustering. Click on “scale and plot”. After results are calculated, differences between samples are displayed inside a non-metric multidimensional scaling plot. Brush points to see which point belongs to which sample. The average silhouette coefficient is plotted below for each potential number of clusters your data can be partitioned into. The higher the value the better the partitioning into clusters between samples. This is also the number of colors used in the network plot, showing which samples are more closely related to each other and build a potential cluster. The tables below use the Indicator species Index to determine which molecular formulae are the strongest representatives for their respective cluster. For details and references see publication.
Figure 15: Statistical comparison of multiple samples with ICBM-OCEAN using non-metric multidimensional scaling (NMDS) based on Bray-Curtis dissimilarity. The upper panel visualizes the probabilities of the sample set being separated into a variable number of groups with the highest “average silhouette value” being the most plausible. The bottom panel visualizes the subgroup definition within the sample set.
3.4 Recommended profile of settings
Additional isotope ratio tolerance: 1000
Tolerance (ppm): 0.5
Elements: C (1-100), H (1-200), O (0-50), N (0-6), S (0-2), P (0-1)
Homologous series: CH2 and (O or H2O or H2)
NSP rule = selected
Either deselect singlets or use the slider “Exclude formulae with intensities present below chosen percentage of samples:” to delete singlets.
Notes when starting from Bruker software:
If you use Bruker software to export your mass lists, we supply the following VBA script that can be used to automatically export your .csv files from Bruker into the format needed for ICBM-OCEAN. Make sure you create the folders “Export from Data Analysis” and “masslists”.
For more info contact: icbm-ocean@uni-oldenburg.de
Analysis.Spectra.MassListClear
Analysis.Spectra.MassListFind 90, 1500
Dim name
name = "D:\Data\Export from Data Analysis\masslists" & CStr(Analysis.Name)
Analysis.Spectra(1).ExportMassList name, daCSV



